Tessa Charles describes the impressive progress being made by synchrotron X-ray facilities to solve the structure of SARS-CoV-2.
At a time when many countries are locking down borders, limiting public gatherings, and encouraging isolation, the Diamond Light Source in Oxfordshire, UK, has been ramping up its intensity, albeit in an organised and controlled manner. The reason: these scientists are working tirelessly on drug-discovery efforts to quell COVID-19.
It is a story that requires fast detectors, reliable robotics and powerful computing infrastructures, artificial intelligence, and one of the brightest X-ray sources in the world. And it is made possible by international collaboration, dedication, determination and perseverance.
Synchrotron light sources are particle accelerators capable of producing incredibly bright X-rays, by forcing relativistic electrons to accelerate on curved trajectories. Around 50 facilities exist worldwide, enabling studies over a vast range of topics. Fanning out tangentially from Diamond’s 562-m circumference storage ring are more than 30 beamlines equipped with instrumentation to serve a multitude of user experiments. The intensely bright X-rays (corresponding to flux of around 9 × 1012 photons per second) are necessary for determining the atomic structure of proteins, including the proteins which make up viruses. As such, synchrotron light sources around the world are interrupting their usual operations to work on mapping the structure of the SARS-CoV-2 virus.
Knowing the atomic structure of the virus is like knowing how the enemy thinks
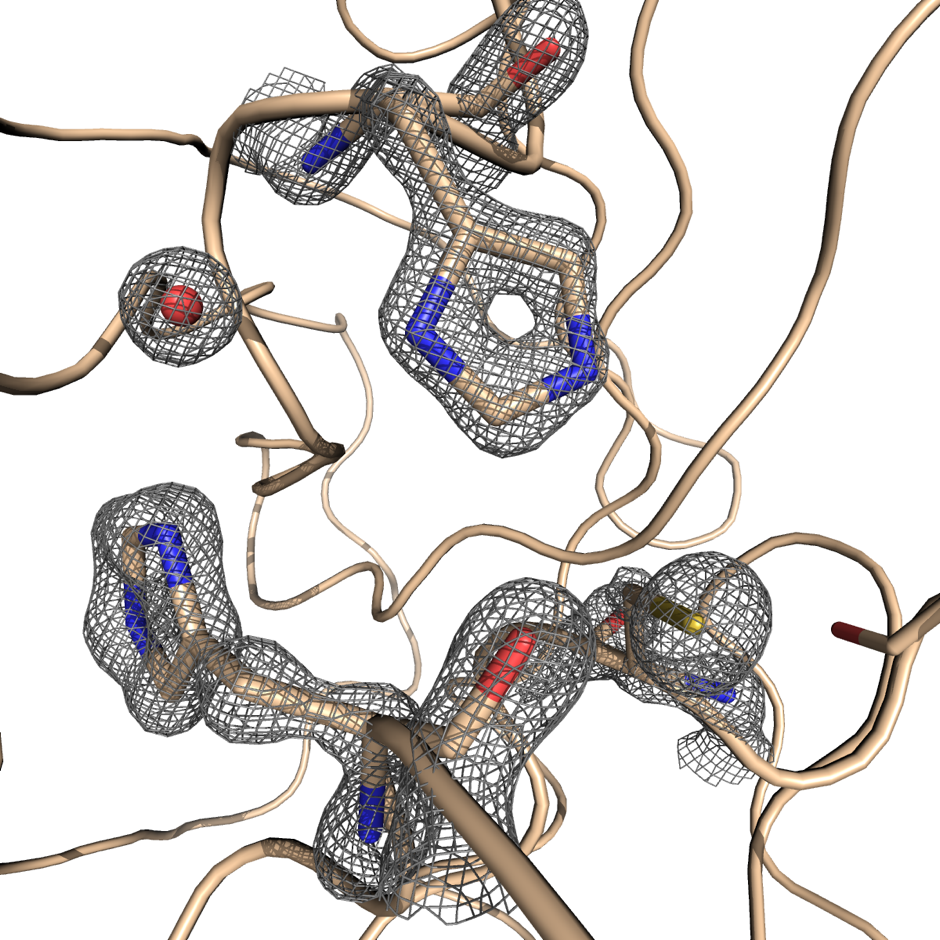
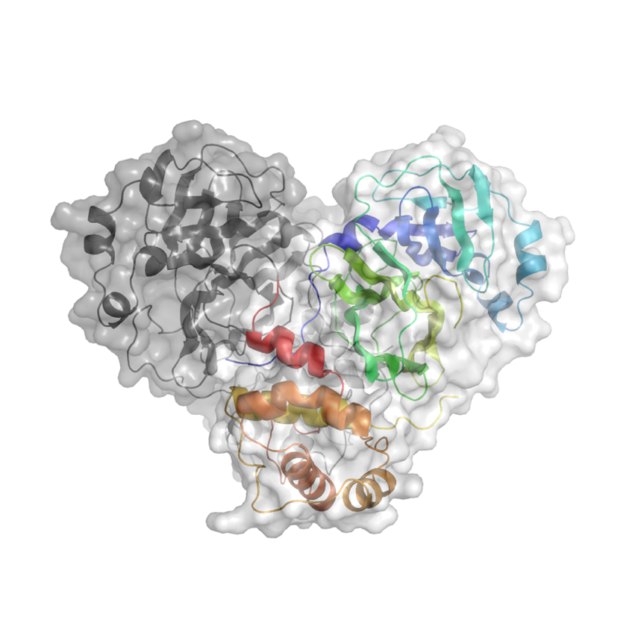
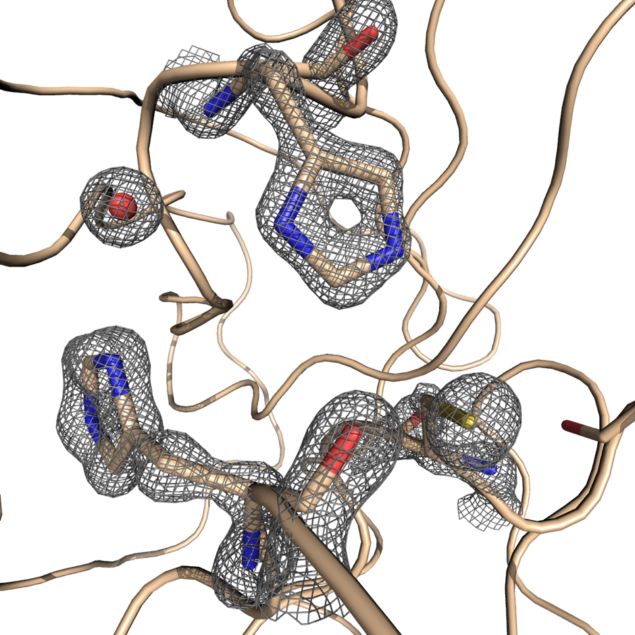
Knowing the atomic structure of the virus is like knowing how the enemy thinks. A 3D visualisation of the building blocks of the structure at an atomic level would allow scientists to understand how the virus functions. Enzymes, the molecular machines that allow the virus to replicate, are key to this process. Scientists at Diamond are exploring the binding site of the main SARS-CoV-2 protease. A drug that binds to this enzyme’s active site would throw a chemical spanner in the works, blocking the virus’ ability to replicate and limiting the spread of the disease.
By way of reminder: Coronavirus is the family of viruses responsible for the common cold, MERS, SARS, etc. Novel coronavirus, aka SARS-CoV-2, is the newly discovered type of coronavirus, and COVID-19 is the disease which it causes.
Call to arms
On 26 January, Diamond’s life-sciences director, Dave Stuart, received a phone call from structural biologist Zihe Rao of ShanghaiTech University in China. Rao, along with his colleague Haitao Yang, had solved the structure of the main SARS-CoV-2 protease with a covalent inhibitor using the Shanghai Synchrotron Radiation Facility (SSRF) in China. Furthermore, they had made the solution freely and publicly available on the worldwide Protein Data Bank.
During the phone call, Rao informed Stuart that their work had been halted by a scheduled shutdown of the SSRF. The Diamond team rapidly mobilised. Since shipping biological samples from Shanghai at the height of the coronavirus in China was expected to be problematic, the team at Diamond ordered the synthetic gene. A synthetic gene can be generated provided the ordering of T, A, C and G nucleotides in the DNA sequence is known. That synthetic gene can be genetically engineered into a bacterium, in this case Escherichia. coli, which reads the sequence and generates the coronavirus protease in large enough quantities for the researchers at Diamond to determine its structure and screen for potential inhibitors.
Eleven days later on 10 February, the synthetic gene arrived. At this point, Martin Walsh, Diamond’s deputy director of life sciences, and his team (consisting of Claire Strain-Damerell, Petra Lukacik, and David Owen) dropped everything. With the gene in hand, the group immediately set up experimental trials to try to generate protein crystals. In order to determine the atomic structure, they needed a crystal containing millions of proteins in an ordered grid-like structure.

X-ray radiation bright enough for the rapid analysis of protein structures can only be produced by a synchrotron light source. The X-rays are directed and focused down a beamline onto a crystal and, as they pass through it, they diffract. From the diffraction pattern, researchers can work backwards to determine the 3D electron density maps and the structure of the protein. The result is a complex curled ribbon-like structure with an intricate mess of twists and turns of the protein chain.
The Diamond team set up numerous trials trying to find the optimum conditions for crystallization of the SARS-CoV-2 protease to occur. They modified the pH, the precipitating compounds, chemical composition, protein to solution ratio… every parameter they could vary, they did. Every day they would produce a few thousand trials, of which only a few hundred would produce crystals, and even fewer would produce crystals of sufficient quality. Within a few days of receiving the gene, the first crystals were being produced. They were paltry and thin crystals but large enough to be tested on one of Diamond’s macromolecular crystallography beamlines.
Watching the results come through, Diamond postdoc David Owen described it as the first moment of intense excitement. With crystals that appeared to be “flat like a car wind shield,” he was dubious as to whether they would diffract at all. Nevertheless, the team placed the crystals in the beamline with a resignation that quickly turned into intense curiosity as the results started appearing before them. At that moment Owen remembers his doubts fading, as he thought, “this might just work!” And work it did. In fact, Owen recalls, “they diffracted beautifully.” These first diffraction patterns of the SARS-CoV-2 virus were recorded with a resolution of 1.9 Angstrom (1.9 × 10−10 m) — high enough resolution to see the position of all of the chemical groups that allow the protease to do its work.
By 19 February, through constant adjustments and learning, the team knew they could grow good-quality crystals quickly. It was time to bring in more colleagues. The XChem team at Diamond joined the mission to set up fragment-based screening – whereby a vast library of small molecules (“fragments”) are soaked into crystals of the viral protease. These fragments are significantly smaller and functionally simpler than most drug molecules and are a powerful approach to selecting candidates for early drug discovery. By 26 February, 600 crystals had been mounted and the first fragment screen launched. In parallel, the team had been making a series of sample to send to company in Oxford called Exscientia, which has set up an AI platform designed to expediate candidates in drug discovery.
Drug-discovery potential
As of early March, 1500 crystals and fragments have been analysed. Owen attributes the team’s success so far to the incredible amounts of data they could collect and analyse quickly. With huge numbers of data sets, they could pin down the parameters of the viral protease with a high degree of confidence. And with the synchrotron light source they were able to create and analyse the diffraction patterns rapidly. The same amount of data collected with a lab-based X-ray source would have taken approximately 10 years. At Diamond, they were able to collect the data in a few days of accumulated beamtime.
Synchrotron light sources all over the world have been granting priority and rapid access to researchers to support their efforts in discovering more about the virus. Researchers at the Advanced Photon Source in Argonne, US, and at Elettra Sincrotrone in Trieste, Italy are also trying to identify molecules effective against COVID-19, in an attempt to bring us closer to an effective vaccine or treatment. This week, the ESRF in Grenoble, France, announced that it will make its cryo-electron microscope facility available for use. The community has a platform called www.lightsources.org offering an overview of access and calls for proposals.
Synchrotron light sources all over the world have been granting priority and rapid access
In addition to allowing the structure of tens of thousands of biological structures to be elucidated – such as that of the ribosome, which was recognised by the 2009 Nobel Prize in Chemistry — light sources have a strong pedigree in elucidating the structure of viruses. Development of common anti-viral medication that blocks the actions of virus in the body, such as Tamiflu or Relenza, also relied upon synchrotrons to reveal their atomic structure.
Mapping the SARS-CoV-2 protease structures bound to small chemical fragments, the Diamond team demonstrated a crystallography- and fragmentation-screen tour de force. The resulting and ongoing work is a crucial first step in developing a drug. Forgoing the usual academic root of peer-review, the Diamond team have made all of their results openly and freely available to help inform public heath response, limit the spread of the virus with the hope that this can fast-track effective treatment options.